Projects
Cannonical GTPase Cycle
Our lab studies Ras and related small GTPases. The canonical signaling cycle involves the transition from the GTP-bound state that actively propagates a variety of signaling cascades by binding effector proteins and the inactive GDP-bound state. GTPase activating proteins (GAPs) bind the GTP-bound form and increase the rate of GTP hydrolysis to end signaling, while guanine nucleotide-exchange factors (GEFs) bind the GDP-bound state prying open the active site to reset the signal.
We have proposed a novel mechanism for hydrolysis in the absence of GAPs in which Raf kinase binds to Ras and facilitates the dimerization of Ras through helixes four and five. In this dimeric state, Ras is primed for allosteric modulation of switch II.
Dimerization & Allosteric Networks
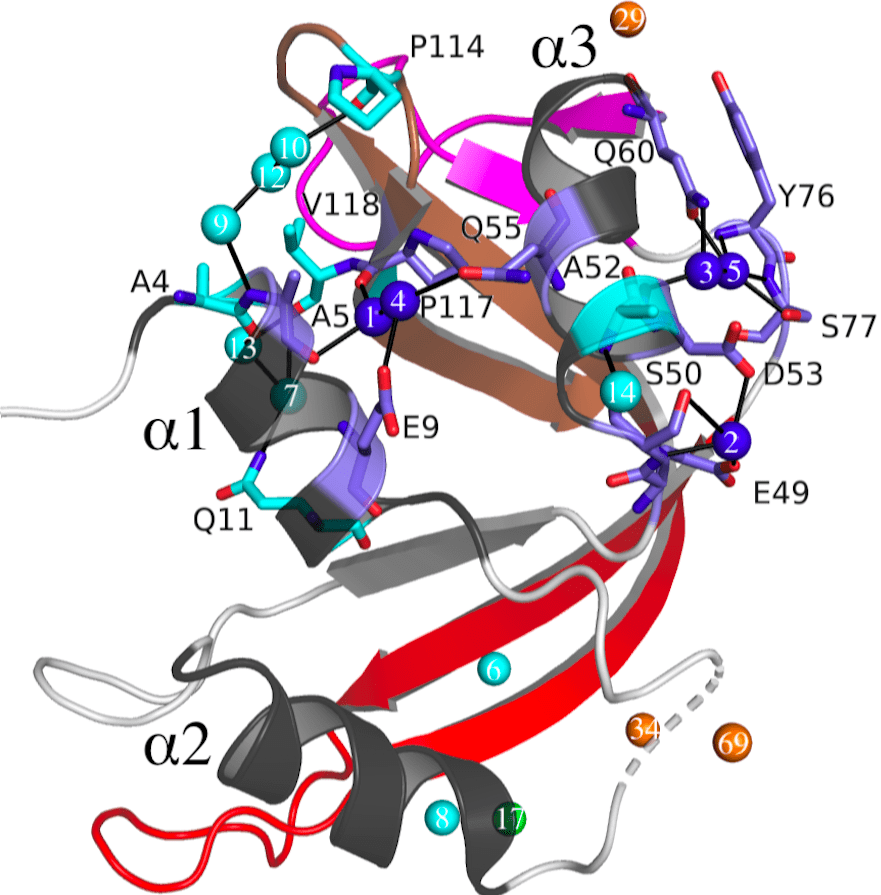
Our lab performs molecular dynamics simulations to explore the protein conformational landscape and understand the allosteric paths connecting distant parts of the protein surface. Network analysis is applied to alpha carbon of the protein backbone serving as a “node”, while “edges” are the lines connecting residues that are at least 4.5 Å apart for 70% or more of the simulation time. We can group these residues into “communities” of residues that interact more closely with each other. Our data indicates the binding of Raf kinase, and subsequent Ras dimerization, increases allosteric connections across the complex. Learn more here.
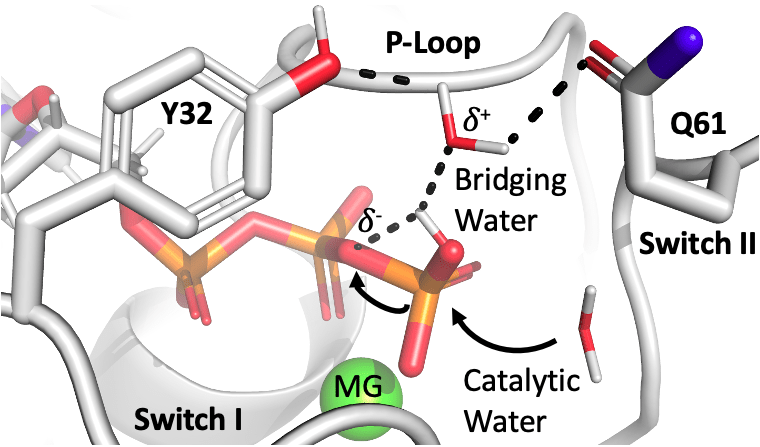
Ras Active Site & GTPase Hydrolysis
Active Site Conformational States
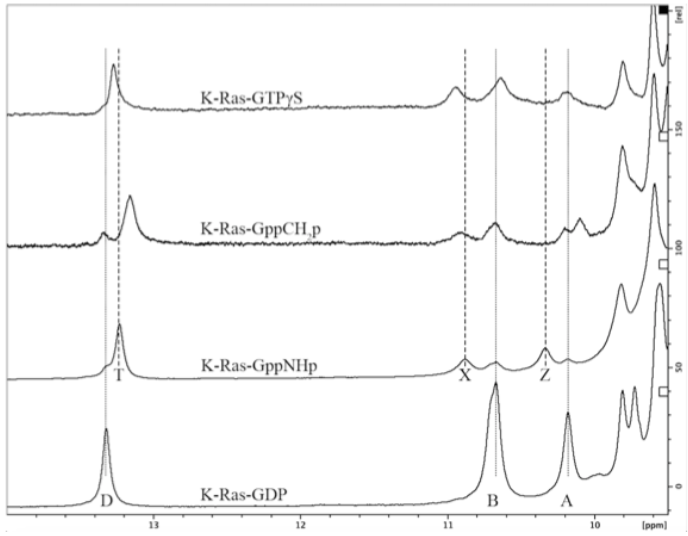
In collaboration with Spiro Pavlopoulos, our lab has discovered downfield signatures sensitive to changes in the active site. We have unambiguously assigned the furthest downfield peak as the proton on N1 of the guanine base; this peak corresponds to either the GTP-bound (“T”) state or the GDP-bound (“D”) state. Learn more here .
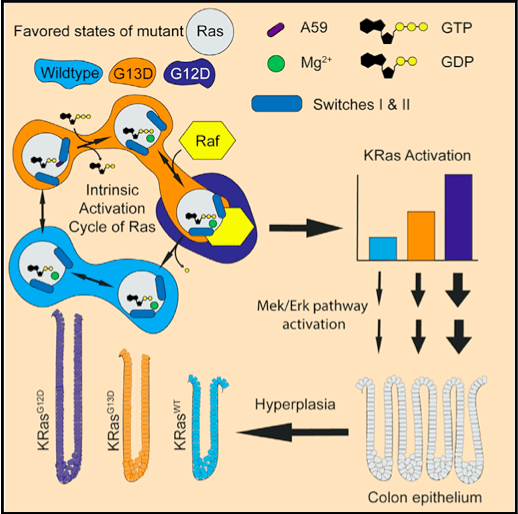
Ras in Human Diseases
Ras signaling pathways play an essential role in controlling cell growth, survival, and differention. Mutations in the Ras gene often results in oncogenesis (somatic mutations) or a class of syndromes termed “Rasopathies” (germline mutations). Approximately 20% of all human cancers contain a mutant copy of the Ras genome. Positions 12, 13, and 61, which surround the active site have the highest frequency of mutation with different levels of occurence between isoforms (Harvey Rat sarcoma “HRas”, Kirsten rat sarcoma “KRas”, Neuroblastoma RAS, “NRas”). For example, mutations at position 12 occur frequntly in KRas, while mutations at position 61 are common in NRas. Our lab is interested in understanding the effects these variants have on the structure through biochemical and biophysical characterization. Learn more here.
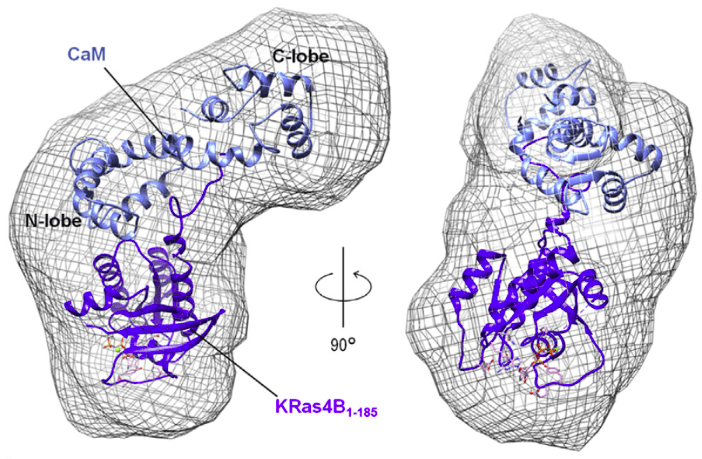
Characterizing Ras-Effector Interactions
Effector binding to GTP-bound Ras and subsequent signal activation is key to many essential biological processes. As Ras is about to interact with many different effector proteins at largely the same Switch I and Switch II regions, our lab investigates the thermodynamic properties of the interaction. We utilize a holistic approach consisting of x-ray crystallography, x-ray solution scattering (SAXS/WAXS), NMR, and molecular dynamics to model these interactions at the atomic level. Learn more here.
DRoP & MSCS
X-Ray Crystallography
The X-ray facilities include two crystal growth chambers (18 and 4 degrees C) outfitted with vibration-free platforms.